|
|
 |
 |
 |
|
|
 |
|
Docking Success Rate
The docking success rate was benchmarked as a percentage of correctly docked ligands
for which the top-scored pose was within 2 Å RMSD from the reference ligand coordinates,
for a set of protein-ligand complexes extracted from PDB. Although alternative definitions
of docking success rate are available1 we used the most widely accepted one in order to make Lead Finder
benchmarks comparable with competitive software benchmarks obtained elsewhere. Additionally, to increase
reproducibility all docking calculations were performed independently 20 times, and
docking was considered successful only
when the probability of generating the top-ranked pose with 2 Å or better accuracy was
at least 0.5.
A set of 407 protein-ligand complexes was used for current docking success rate measurements.
This set of complexes was combined from test sets used in original benchmarking studies of such
docking software as: FlexX2,
Glide SP3,
Glide XP4,
Gold5,6,7,
LigandFit8,
MolDock9,
Surflex10.
For this reason, current benchmarking study of Lead Finder docking success rate
appears to be the most extensive study of such kind; moreover, the results obtained with Lead Finder
can be correctly compared to competitive docking programs of interest
since they were obtained on the same sets of protein-ligand complexes.
All benchmarking calculations were performed in two regimes
(docking and screening, see Docking Algorithm section).
The docking success rate obtained with Lead Finder
on different test sets ranged from 80.0% (for GlideXP and FlexX test sets) to 96.0% (for Surflex
and MolDock test sets). The data in the table below show that Lead Finder outperformed
all reviewed docking software programs for which reliable original benchmarks were available.
More specifically, Lead Finder
successfully docked 73 structures which could not be docked by FlexX,
52 structures - for Glide SP, 52 structures - for
Glide XP, 30 structures - for Gold on the original Gold test set,
13 structures - for Gold on Astex diversity test set, 9 structures -
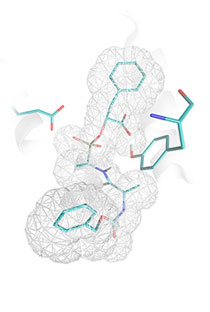
for MolDock, 21 structures - for Surflex. Example of carboxypeptidase A inhibitor,
which could not be correctly docked by such programs
as Glide3
or FlexX2,
but was successfully docked by Lead Finder, is illustrated in the figure.
 |
|
a) Data for the high-resolution subset (92 structures with resolution better than 2 Å) are not provided in original publication, only overall performance (over entire CCDC/Astex test of 224 structures) is given.
b) Original publication contains data only for 19 complexes, which is too small for comparative purposes. We extended this set by including additional 75 protein-ligand complexes which were used in original publication to benchmark LigandFit ability to detect active site cavity.  |
|
|
The docking success rate achieved by Lead Finder in the faster, less accurate screening regime
was also higher than the original benchmarks obtained with
competitive software, except MolDock. Such results were achieved by adjusting settings
of the docking algorithm to minimize decrease in docking success rate
while increasing the speed of calculations.
The applicability of screening regime for high-performance calculations is illustrated
in the Computing Speed section.
 |
|
|
E-mail: info@biomoltech.com |
Phone: +1(416)238-1263 |
Fax: +1(416)352-6117 Mailing address: 226 York Mills Rd, Toronto, Ontario M2L 1L1, Canada |