|
Benchmarking Lead Finder's Performance
The quality of Lead Finder predictions was extensively benchmarked in a number of computational experiments:
- Virtual Screening Performance
The ability of Lead Finder
to distinguish active compounds from inactive ones during virtual screening experiments has been benchmarked
against 34 therapeutically relevant protein targets. For almost all targets
Lead Finder demonstrated significant enrichment indicators, confirming its applicability
for in silico lead compound discovery.
- Docking Success Rate
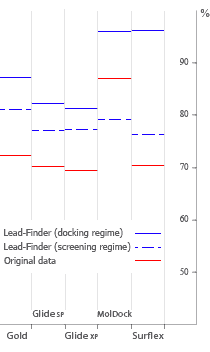
The accuracy of protein-ligand docking has been assessed on a set of 407 structures
that comprises practically all published test sets of such software applications as
FlexX,
Glide SP and Glide XP,
Gold,
LigandFit,
MolDock,
Surflex.
Lead Finder successfully predicted structures for
85% of protein-ligand complexes, notably exceeding the results demonstrated by
other docking software.
- Binding Energy Prediction
The ability of Lead Finder to estimate free energy of protein-ligand
binding has been benchmarked against a set of 330 diverse protein-ligand
complexes. Lead Finder demonstrated high accuracy of binding energy prediction with
RMSD of 1.5 kcal/mol)
at high speed of calculations (less than one second per compound on average on a standard single-processor Pentium PC).
- Computing Speed
To estimate time and computational resources necessary for your computational tasks, the speed of ligand
docking on a single processor/core has been benchmarked on different compounds
with varying number of freely rotatable bonds, computing environments, and configuration settings
of the docking algorithm.
 |
|
Docking success rate (%) of different programs obtained on their native test sets and current Lead Finder benchmarks in default docking and screening regimes.
|
|
|